Gene Ontology overview
The Gene Ontology (GO) is a structured, standardized representation of biological knowledge. GO describes concepts (also known as terms, or formally, classes) that are connected to each other via formally defined relations. The GO is designed to be species-agnostic to enable the annotation of gene products across the entire tree of life. The computational framework of the GO enables consistent gene annotation, comparison of functions across organisms, and integration of knowledge across diverse biological databases.
GO aspects
The GO is organized in three aspects: Molecular Function (MF), Cellular Component (CC), and Biological Process (BP).
Molecular Function
MFs represent molecular-level activities performed by gene products, such as “catalysis” or “transcription regulator activity”. MFs correspond to activities that can be performed by individual gene products (i.e. a protein or RNA), but some activities are performed by molecular complexes composed of multiple gene products, when the activity cannot be ascribed to a single gene product of the complex. Examples of broad functional terms are catalytic activity and transporter activity; examples of more specific functional terms are adenylate cyclase activity or insulin receptor activity.
GO MF terms represent activities and not the entities that perform the actions. To avoid confusion between gene product names and their molecular activities, GO MFs are appended with the word “activity” (a protein kinase would have the GO MF protein kinase activity). Finally, MFs do not specify where, when, or in what context the action takes place.
Cellular Component
CC serves to capture the cellular location where a molecular function takes place. CCs include:
- cellular anatomical structures, encompassing cellular entities such as the plasma membrane and the cytoskeleton, as well as membrane-enclosed cellular compartments such as the mitochondrion.
- stable protein-containing complexes of which they are parts.
- virion components, classified separately because viruses are not cellular organisms. Examples include viral capsid and viral envelope.
Biological Process
BPs are the larger processes or ‘biological programs’ accomplished by the concerted action of multiple molecular activities. Examples of broad BP terms are DNA repair or signal transduction. Examples of more specific terms are cytosine biosynthetic process or D-glucose transmembrane transport.
Root Terms
Each of the three GO aspects is represented by a separate root ontology term. Moreover, the three GO aspects are is a disjoint, meaning that no is a relation exists between terms from the different ontology aspects. However, other relationships such as part of and occurs in can operate between terms from different GO aspects. For example, the MF term cyclin-dependent protein kinase activity is part of the BP regulation of cell cycle.
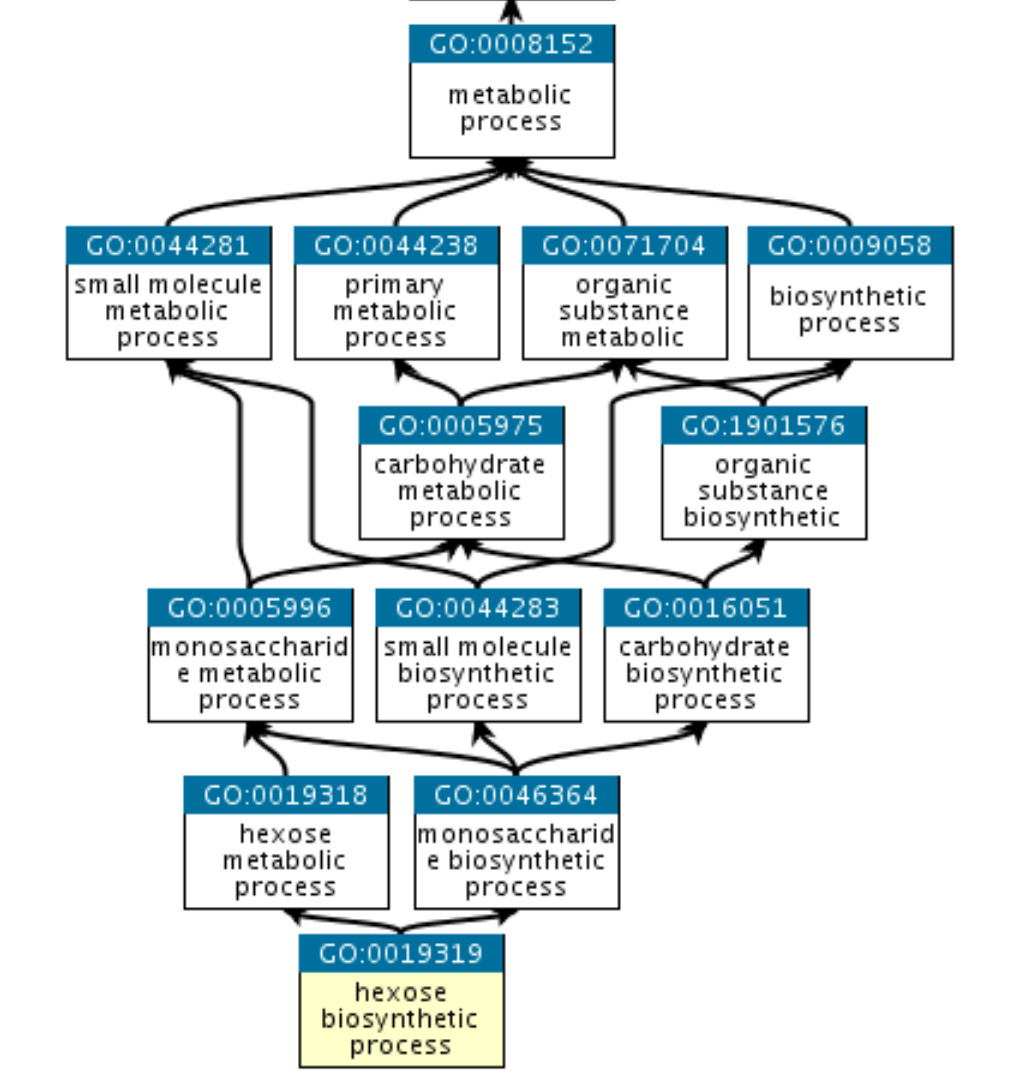
The GO hierarchy
The GO is structured as a graph in which each GO term is a node and the relationships between the nodes are edges. GO is hierarchical, with child terms being more specialized than their parent terms, but unlike a strict hierarchy, a term may have more than one parent term (note that the parent/child model does not hold true for all types of relations, see the relations documentation). For example, the biological process term hexose biosynthetic process has two parents, hexose metabolic process and monosaccharide biosynthetic process. This reflects the fact that biosynthetic process is a subtype of metabolic process, and a hexose is a subtype of monosaccharide.
GO term elements
The different elements of a GO term are show in the image below (screen shot from the AmiGO GO browser).
Mandatory elements
- Accession (also known as Unique identifier): Every term has a GO ID, a unique seven digit identifier prefixed by GO:, e.g. GO:0005739, GO:1904659, or GO:0016597.
- Term name: Every term has a human-readable term name — e.g. mitochondrion, D-glucose transmembrane transport, or amino acid binding.
- Ontology (also known as Aspect): Denotes which of the three sub-ontologies the term belongs to. Written as molecular_function (MF), biological_process (BP) and cellular_component (CC).
- Definition: A textual description of what the term represents, plus reference(s) to the source of the information.
- Relationships to other terms: How the term relates to other terms in the ontology. All terms (other than the root terms representing each aspect, above) have an is a sub-class relationship to another term. The Gene Ontology employs a number of other relations; the relations documentation page describes the relations used in the ontology.
Optional elements
- Alternate ID (also known as Secondary IDs): Secondary IDs come about when two or more terms are identical in meaning, and are merged into a single term. All terms IDs are preserved so that no information (for example, annotations to the merged IDs) is lost.
- Synonyms: Alternative words or phrases closely related in meaning to the term name, with indication of the relationship between the name and synonym given by the synonym scope. The scopes for GO synonyms are:
- Exact: an exact equivalent; interchangeable with the term name; for e.g. ornithine cycle is an exact synonym of *urea cycle *
- Broad: the synonym is broader than the term name; for e.g. cell division is a broad synonym of cytokinesis
- Narrow: the synonym is narrower or more precise than the term name; for e.g. pyrimidine-dimer repair by photolyase is a narrow synonym of photoreactive repair
- Related: the terms are related in some imprecise way; for e.g. cytochrome bc1 complex is a related synonym of ubiquinol-cytochrome-c reductase activity; virulence is a related synonym of pathogenesis.
- Custom synonym types are also used in the ontology. For example, a number of synonyms are designated as systematic synonyms; synonyms of this type are exact synonyms of the term name.
- Comment: Any extra information about the term and its usage.
- Chem. react.: For terms having cross references to the RHEA database of chemical reactions, this section lists the reaction paticipants.
- Subset: Indicates that the term belongs to one or more GO subsets.
- Obsolete tag: Boolean value that indicates that the term has been deprecated and should not be used. A GO term is obsoleted when it is out of scope, misleadingly named or defined, or describes a concept that would be better represented in another way and needs to be removed from the published ontology. In these cases, the term and ID still persist in the ontology, but the term is tagged as obsolete, and all relationships to other terms are removed. A comment is added to the term detailing the reason for the obsoletion and replacement terms are suggested whenever possible.
- Taxon constraints: Annotations to some GO terms are restricted to specific species; the taxon constraints specify which taxa a term can be applied to.
- Database cross-references: Database cross-references, or dbxrefs, refer to identical or very similar objects in other databases. For instance, the molecular function term cytosine deaminase activity is cross-referenced to RHEA:20605; the biological process term sulfate assimilation has the InterPro cross-reference Sulphate adenylyltransferase (IPR002650). Database cross-references are visible from the tab at the bottom of the term description (as shown in screenshot below).
GO is a dynamic ontology
GO aims to represent the current state of knowledge in biology, hence it is constantly revised and expanded as biological knowledge accumulates. Revisions to the ontology are managed by a team of editors with broad biological knowledge and expertise in computational knowledge representation. GO updates are made collaboratively between the GOC ontology team and scientists who request the updates. Most requests come from scientists making GO annotations (these typically impact only a few terms each), and from domain experts in particular areas of biology (these typically revise an entire ‘branch’ of the ontology comprising many terms and relations). Changes to the ontology can be visualized on the GO statistics page. We welcome researchers and computational scientists to submit requests for either new terms, new relations, or any other improvements to the ontology.